A mere 1% of our genome is dedicated to protein-coding sequences, while a far greater fraction of our A’s, T’s, C’s and G’s encode regulatory information – and over 90% of disease-associated variants disrupt these regulatory sequences. Deciphering the regulatory genome and uncovering the molecular mechanisms of gene regulation is therefore essential to our understanding of development, physiology and disease.
Strikingly, many important genes are controlled by constellations of enhancers scattered over vast regulatory landscapes, and active in different cellular contexts. Spatial genome organization is therefore thought to shape the timing and specificity of these long-range interactions. We recently showed that two distinct classes of architectural elements (one that promotes specific interactions, another that precludes spurious ones) control the expression of key developmental regulators, the Hox genes (Batut et al., 2022).
What are the sequence elements and protein factors that establish genome architecture? And how is it leveraged to regulate gene expression?
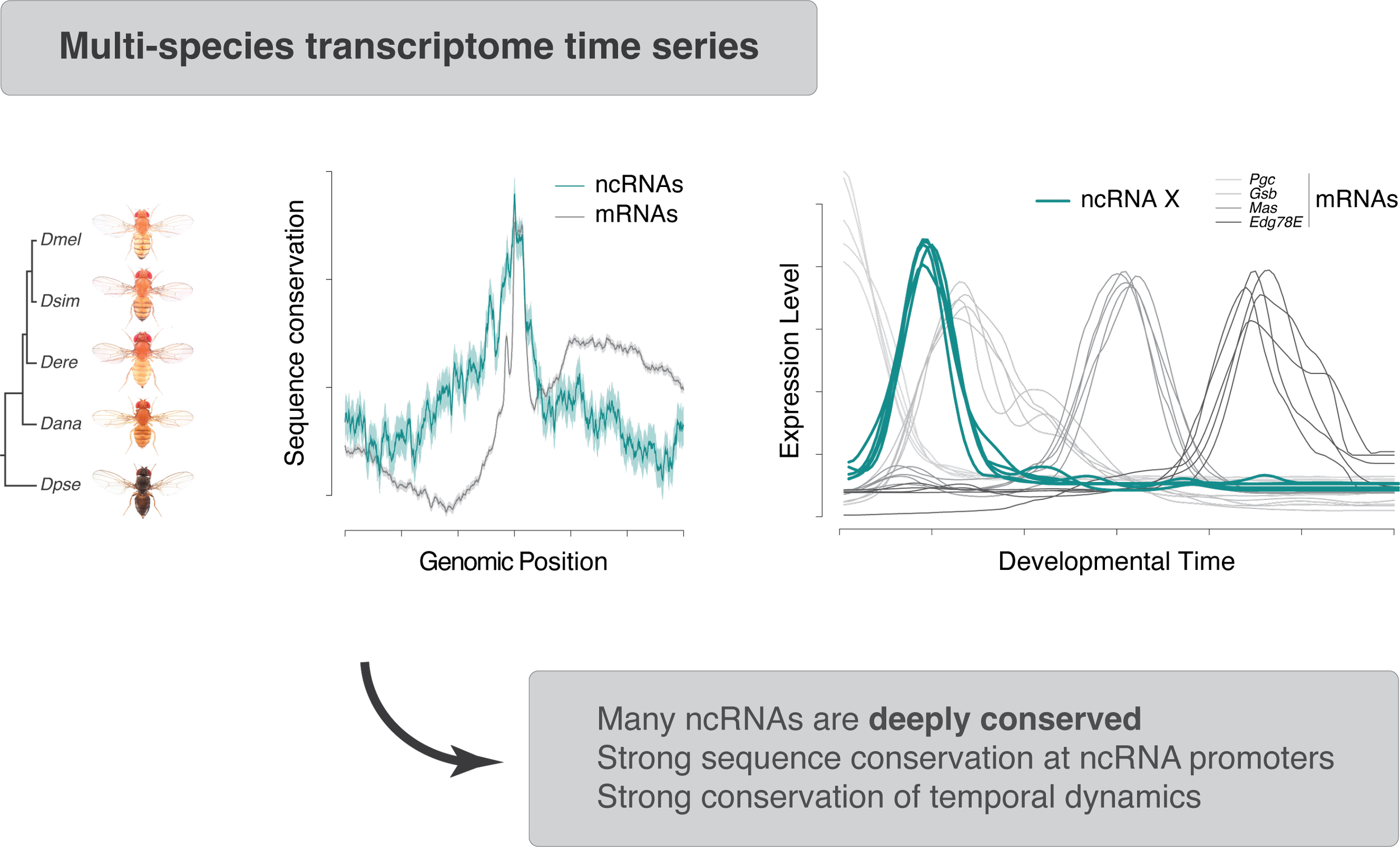
Many enhancers and architectural elements are also transcribed into noncoding (nc)RNAs. Although we have long known that ncRNAs are at the heart of many fundamental processes – from mRNA splicing to protein synthesis and telomere maintenance – the sheer scale and pervasiveness of noncoding transcription was a transformative insight of the post-genome era, including our efforts as part of the ENCODE Project (Djebali et al., 2012). Do these ncRNAs matter? We used evolutionary transcriptomics to show that hundreds of ncRNAs transcribed in Drosophila embryos are deeply conserved, and therefore probably functional (Batut & Gingeras, 2017).
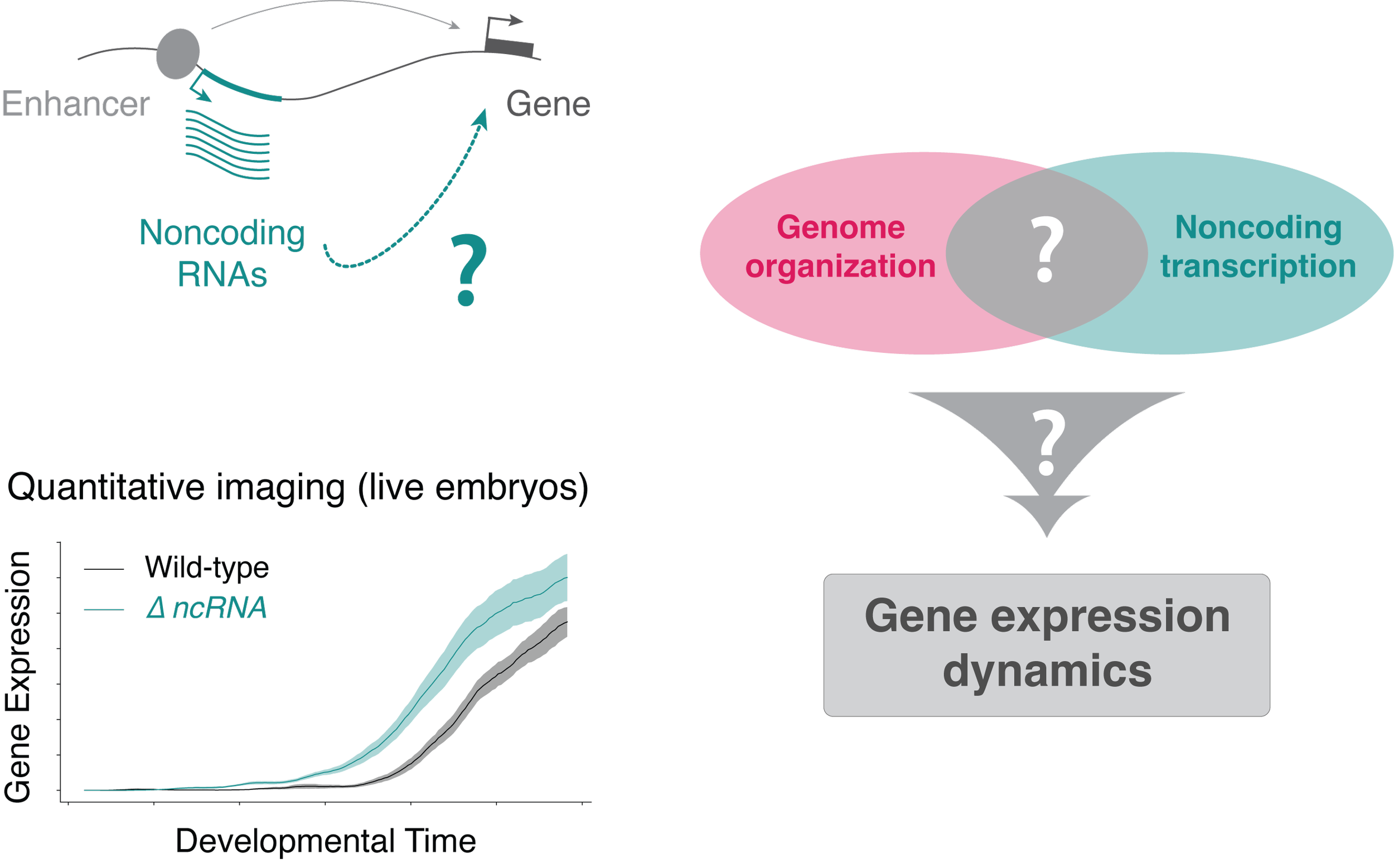
The observation that so much noncoding transcription emanates from regulatory elements suggests that it might play a global role in controlling gene expression. Our lab is particularly interested in how ncRNAs might both exploit and directly control aspects of genome organization to shape regulatory outcomes.
Does noncoding transcription regulate gene expression? Through what molecular mechanisms? And what is the interplay between ncRNAs and 3D genome organization?
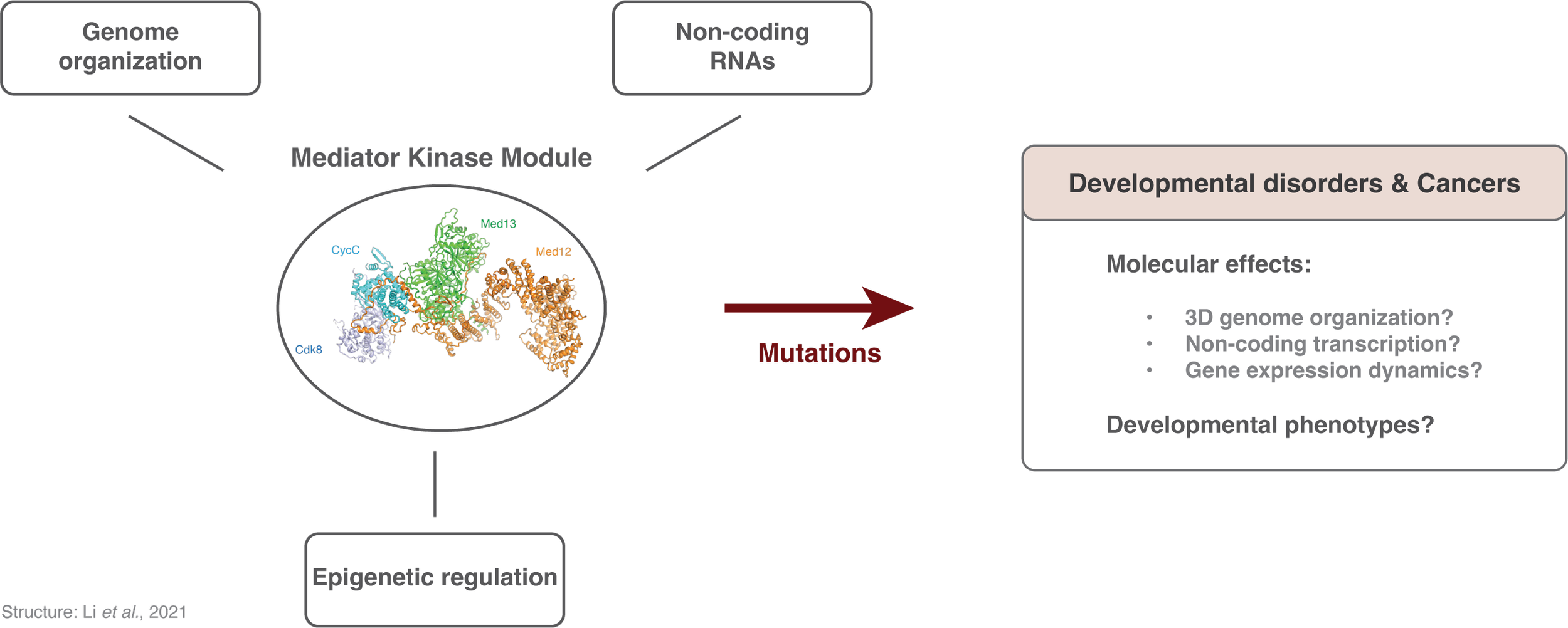
Gene expression patterns initially established by the binding of transcription factors to enhancers must often be maintained throughout developmental trajectories, and sometimes over the lifetime of the organism. Epigenetic mechanisms involving the Polycomb and Trithorax systems play a role in the maintenance of active or repressed chromatin states.
How do noncoding transcription and genome organization control these epigenetic systems? And how are these different regulatory modalities integrated together?
Our lab studies these questions in the exquisitely dynamic context of embryonic development, using single-cell live imaging approaches to measure the spatiotemporal dynamics of transcription in this highly coordinated regulatory choreography (top). We also directly visualize noncoding transcription in vivo (bottom) to disentangle its complex relationship to gene expression.
These strategies allowed us to reveal that both genome organization (Batut et al., 2022) and noncoding transcription (ongoing work) play a key role in regulating the timing and dynamics of gene expression.
Patterning genes eve & hairy